
Table of Useful R commands
Command Purpose
help() Obtain documentation for a given R command
example() View some examples on the use of a command
c(), scan() Enter data manually to a vector in R
seq() Make arithmetic progression vector
rep() Make vector of repeated values
data() Load (often into a data.frame) built-in dataset
View() View dataset in a spreadsheet-type format
str() Display internal structure of an R object
read.csv(), read.table() Load into a data.frame an existing data file
library(), require() Make available an R add-on package
dim() See dimensions (# of rows/cols) of data.frame
length() Give length of a vector
ls() Lists memory contents
rm() Removes an item from memory
names() Lists names of variables in a data.frame
hist() Command for producing a histogram
histogram() Lattice command for producing a histogram
stem() Make a stem plot
table() List all values of a variable with frequencies
xtabs() Cross-tabulation tables using formulas
mosaicplot() Make a mosaic plot
cut() Groups values of a variable into larger bins
mean(), median() Identify “center” of distribution
by() apply function to a column split by factors
summary() Display 5-number summary and mean
var(), sd() Find variance, sd of values in vector
sum() Add up all values in a vector
quantile() Find the position of a quantile in a dataset
barplot() Produces a bar graph
barchart() Lattice command for producing bar graphs
boxplot() Produces a boxplot
bwplot() Lattice command for producing boxplots
Command Purpose
plot() Produces a scatterplot
xyplot() Lattice command for producing a scatterplot
lm() Determine the least-squares regression line
anova() Analysis of variance (can use on results of lm())
predict() Obtain predicted values from linear model
nls() estimate parameters of a nonlinear model
residuals() gives (observed - predicted) for a model fit to data
sample() take a sample from a vector of data
replicate() repeat some process a set number of times
cumsum() produce running total of values for input vector
ecdf() builds empirical cumulative distribution function
dbinom(), etc. tools for binomial distributions
dpois(), etc. tools for Poisson distributions
pnorm(), etc. tools for normal distributions
qt(), etc. tools for student t distributions
pchisq(), etc. tools for chi-square distributions
binom.test() hypothesis test and confidence interval for 1 proportion
prop.test() inference for 1 proportion using normal approx.
chisq.test() carries out a chi-square test
fisher.test() Fisher test for contingency table
t.test() student t test for inference on population mean
qqnorm(), qqline() tools for checking normality
addmargins() adds marginal sums to an existing table
prop.table() compute proportions from a contingency table
par() query and edit graphical settings
power.t.test() power calculations for 1- and 2-sample t
anova() compute analysis of variance table for fitted model

R Commands for MATH 143 Examples of usage
Examples of usage
help()
help(mean)
example()
require(lattice)
example(histogram)
c(), rep() seq()
> x = c(8, 6, 7, 5, 3, 0, 9)
> x
[1] 8 6 7 5 3 0 9
> names = c("Owen", "Luke", "Anakin", "Leia", "Jacen", "Jaina")
> names
[1] "Owen" "Luke" "Anakin" "Leia" "Jacen" "Jaina"
> heartDeck = c(rep(1, 13), rep(0, 39))
> heartDeck
[1] 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[49] 0 0 0 0
> y = seq(7, 41, 1.5)
> y
[1] 7.0 8.5 10.0 11.5 13.0 14.5 16.0 17.5 19.0 20.5 22.0 23.5 25.0 26.5 28.0 29.5 31.0 32.5 34.0
[20] 35.5 37.0 38.5 40.0
data(), dim(), names(), View(), str()
> data(iris)
> names(iris)
[1] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width" "Species"
> dim(iris)
[1] 150 5
> str(iris)
'data.frame': 150 obs. of 5 variables:
$ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
$ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
$ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
$ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
$ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
> View(iris)
2

R Commands for MATH 143 Examples of usage
ls(), rm()
> data(iris)
> data(faithful)
> data(Puromycin)
> data(LakeHuron)
> ls()
[1] "faithful" "heartDeck" "iris" "LakeHuron" "names" "Puromycin" "x" "y"
> newVector = 1:12
> ls()
[1] "faithful" "heartDeck" "iris" "LakeHuron" "names" "newVector" "Puromycin" "x"
[9] "y"
> rm(faithful)
> ls()
[1] "heartDeck" "iris" "LakeHuron" "names" "newVector" "Puromycin" "x" "y"
hist()
data(faithful)
hist(faithful$eruptions)
hist(faithful$eruptions, n=15)
hist(faithful$eruptions, breaks=seq(1.5,5.25,.25), col="red")
hist(faithful$eruptions, freq=F, n=15, main="Histogram of Old Faithful Eruption Times", xlab="Duration (mins)")
Histogram of Old Faithful Eruption Times
Duration (mins)
Density
1.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0
0.0 0.2 0.4 0.6
3

R Commands for MATH 143 Examples of usage
library(), require()
> library(abd)
> require(lattice)
histogram()
require(lattice)
data(iris)
histogram(iris$Sepal.Length, breaks=seq(4,8,.25))
histogram(~ Sepal.Length, data=iris, main="Iris Sepals", xlab="Length")
histogram(~ Sepal.Length | Species, data=iris, col="red")
histogram(~ Sepal.Length | Species, data=iris, n=15, layout=c(1,3))
read.csv()
> As.in.H2O = read.csv("http://www.calvin.edu/~scofield/data/comma/arsenicInWater.csv")
read.table()
> senate = read.table("http://www.calvin.edu/~scofield/data/tab/rc/senate99.dat", sep="\t", header=T)
mean(), median(), summary(), var(), sd(), quantile(),
> counties=read.csv("http://www.calvin.edu/~stob/data/counties.csv")
> names(counties)
[1] "County" "State" "Population" "HousingUnits" "TotalArea"
[6] "WaterArea" "LandArea" "DensityPop" "DensityHousing"
> x = counties$LandArea
> mean(x, na.rm = T)
[1] 1126.214
> median(x, na.rm = T)
[1] 616.48
> summary(x)
Min. 1st Qu. Median Mean 3rd Qu. Max.
1.99 431.70 616.50 1126.00 923.20 145900.00
> sd(x, na.rm = T)
[1] 3622.453
> var(x, na.rm = T)
[1] 13122165
> quantile(x, probs=seq(0, 1, .2), na.rm=T)
0% 20% 40% 60% 80% 100%
1.99 403.29 554.36 717.94 1043.82 145899.69
4

R Commands for MATH 143 Examples of usage
sum()
> firstTwentyIntegers = 1:20
> sum(firstTwentyIntegers)
[1] 210
> die = 1:6
> manyRolls = sample(die, 100, replace=T)
> sixFreq = sum(manyRolls == 6)
> sixFreq / 100
[1] 0.14
stem()
> monarchs = read.csv("http://www.calvin.edu/~scofield/data/comma/monarchReigns.csv")
> stem(monarchs$years)
The decimal point is 1 digit(s) to the right of the |
0 | 0123566799
1 | 0023333579
2 | 012224455
3 | 355589
4 | 4
5 | 069
6 | 3
table(), table(), mosaicplot(), cut()
> pol = read.csv("http://www.calvin.edu/~stob/data/csbv.csv")
> table(pol$sex)
Female Male
133 88
> table(pol$sex, pol$Political04)
Conservative Far Right Liberal Middle-of-the-road
Female 67 0 14 48
Male 47 7 6 28
> xtabs(~sex, data=pol)
sex
Female Male
133 88
> xtabs(~Political04 + Political07, data=pol)
Political07
Political04 Conservative Far Left Far Right Liberal Middle-of-the-road
Conservative 58 0 2 13 39
Far Right 4 0 3 0 0
Liberal 0 1 1 14 4
Middle-of-the-road 20 0 0 22 32
> mosaicplot(~Political04 + sex, data=pol)
5

R Commands for MATH 143 Examples of usage
pol
Political04
sex
Conservative
Far Right
Liberal
Middle−of−the−road
FemaleMale
> monarchs = read.csv("http://www.calvin.edu/~scofield/data/comma/monarchReigns.csv")
> table(monarchs$years)
0 1 2 3 5 6 7 9 10 12 13 15 17 19 20 21 22 24 25 33 35 38 39 44 50 56 59 63
1 1 1 1 1 2 1 2 2 1 4 1 1 1 1 1 3 2 2 1 3 1 1 1 1 1 1 1
> xtabs(~years, data=monarchs)
years
0 1 2 3 5 6 7 9 10 12 13 15 17 19 20 21 22 24 25 33 35 38 39 44 50 56 59 63
1 1 1 1 1 2 1 2 2 1 4 1 1 1 1 1 3 2 2 1 3 1 1 1 1 1 1 1
> cut(monarchs$years, breaks=seq(0,65,5))
[1] (20,25] (10,15] (30,35] (15,20] (30,35] (5,10] (15,20] (55,60] (30,35] (15,20] (45,50] (20,25]
[13] (10,15] (5,10] (35,40] (20,25] <NA> (0,5] (20,25] (35,40] (5,10] (0,5] (40,45] (20,25]
[25] (20,25] (20,25] (0,5] (10,15] (5,10] (10,15] (10,15] (30,35] (55,60] (5,10] (5,10] (60,65]
[37] (5,10] (20,25] (0,5] (10,15]
13 Levels: (0,5] (5,10] (10,15] (15,20] (20,25] (25,30] (30,35] (35,40] (40,45] (45,50] ... (60,65]
> table(cut(monarchs$years, breaks=seq(0,65,5)))
(0,5] (5,10] (10,15] (15,20] (20,25] (25,30] (30,35] (35,40] (40,45] (45,50] (50,55] (55,60]
4 7 6 3 8 0 4 2 1 1 0 2
(60,65]
1
> fiveYrLevels = cut(monarchs$years, breaks=seq(0,65,5))
> xtabs(~fiveYrLevels)
fiveYrLevels
(0,5] (5,10] (10,15] (15,20] (20,25] (25,30] (30,35] (35,40] (40,45] (45,50] (50,55] (55,60]
4 7 6 3 8 0 4 2 1 1 0 2
(60,65]
1
6

R Commands for MATH 143 Examples of usage
barplot()
pol = read.csv("http://www.calvin.edu/~stob/data/csbv.csv")
barplot(table(pol$Political04), main="Political Leanings, Calvin Freshman 2004")
barplot(table(pol$Political04), horiz=T)
barplot(table(pol$Political04),col=c("red","green","blue","orange"))
barplot(table(pol$Political04),col=c("red","green","blue","orange"),
names=c("Conservative","Far Right","Liberal","Centrist"))
Conservative Liberal Centrist
0 40 80
barplot(xtabs(~sex + Political04, data=pol), legend=c("Female","Male"), beside=T)
Conservative Far Right Liberal Middle−of−the−road
Female
Male
0 10 20 30 40 50 60
7

R Commands for MATH 143 Examples of usage
boxplot()
data(iris)
boxplot(iris$Sepal.Length)
boxplot(iris$Sepal.Length, col="yellow")
boxplot(Sepal.Length ~ Species, data=iris)
boxplot(Sepal.Length ~ Species, data=iris, col="yellow", ylab="Sepal length",main="Iris Sepal Length by Species")
●
setosa versicolor virginica
4.5 5.5 6.5 7.5
Iris Sepal Length by Species
Sepal length
plot()
data(faithful)
plot(waiting~eruptions,data=faithful)
plot(waiting~eruptions,data=faithful,cex=.5)
plot(waiting~eruptions,data=faithful,pch=6)
plot(waiting~eruptions,data=faithful,pch=19)
plot(waiting~eruptions,data=faithful,cex=.5,pch=19,col="blue")
plot(waiting~eruptions, data=faithful, cex=.5, pch=19, col="blue", main="Old Faithful Eruptions",
ylab="Wait time between eruptions", xlab="Duration of eruption")
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
● ●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
● ●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
● ●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
● ●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
1.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0
50 70 90
Old Faithful Eruptions
Duration of eruption
Time between eruptions
8

R Commands for MATH 143 Examples of usage
sample()
> sample(c("Heads","Tails"), size=1)
[1] "Heads"
> sample(c("Heads","Tails"), size=10, replace=T)
[1] "Heads" "Heads" "Heads" "Tails" "Tails" "Tails" "Tails" "Tails" "Tails" "Heads"
> sample(c(0, 1), 10, replace=T)
[1] 1 0 0 1 1 0 0 1 0 0
> sum(sample(1:6, 2, replace=T))
[1] 10
> sample(c(0, 1), prob=c(.25,.75), size=10, replace=T)
[1] 1 1 1 0 1 1 1 1 1 1
> sample(c(rep(1,13),rep(0,39)), size=5, replace=F)
[1] 0 0 0 0 0
replicate()
> sample(c("Heads","Tails"), 2, replace=T)
[1] "Tails" "Heads"
> replicate(5, sample(c("Heads","Tails"), 2, replace=T))
[,1] [,2] [,3] [,4] [,5]
[1,] "Heads" "Tails" "Heads" "Tails" "Heads"
[2,] "Heads" "Tails" "Heads" "Heads" "Heads"
> ftCount = replicate(100000, sum(sample(c(0, 1), 10, rep=T, prob=c(.6, .4))))
> hist(ftCount, freq=F, breaks=-0.5:10.5, xlab="Free throws made out of 10 attempts",
+ main="Simulated Sampling Dist. for 40% FT Shooter", col="green")
Simulated Sampling Dist. for 40% FT Shooter
Free throws made out of 10 attempts
Density
0 2 4 6 8 10
0.00 0.15
9

R Commands for MATH 143 Examples of usage
dbinom(), pbinom(), qbinom(), rbinom(), binom.test(), prop.test()
> dbinom(0, 5, .5) # probability of 0 heads in 5 flips
[1] 0.03125
> dbinom(0:5, 5, .5) # full probability dist. for 5 flips
[1] 0.03125 0.15625 0.31250 0.31250 0.15625 0.03125
> sum(dbinom(0:2, 5, .5)) # probability of 2 or fewer heads in 5 flips
[1] 0.5
> pbinom(2, 5, .5) # same as last line
[1] 0.5
> flip5 = replicate(10000, sum(sample(c("H","T"), 5, rep=T)=="H"))
> table(flip5) / 10000 # distribution (simulated) of count of heads in 5 flips
flip5
0 1 2 3 4 5
0.0310 0.1545 0.3117 0.3166 0.1566 0.0296
> table(rbinom(10000, 5, .5)) / 10000 # shorter version of previous 2 lines
0 1 2 3 4 5
0.0304 0.1587 0.3087 0.3075 0.1634 0.0313
> qbinom(seq(0,1,.2), 50, .2) # approx. 0/.2/.4/.6/.8/1-quantiles in Binom(50,.2) distribution
[1] 0 8 9 11 12 50
> binom.test(29, 200, .21) # inference on sample with 29 successes in 200 trials
Exact binomial test
data: 29 and 200
number of successes = 29, number of trials = 200, p-value = 0.02374
alternative hypothesis: true probability of success is not equal to 0.21
95 percent confidence interval:
0.09930862 0.20156150
sample estimates:
probability of success
0.145
> prop.test(29, 200, .21) # inference on same sample, using normal approx. to binomial
1-sample proportions test with continuity correction
data: 29 out of 200, null probability 0.21
X-squared = 4.7092, df = 1, p-value = 0.03
alternative hypothesis: true p is not equal to 0.21
95 percent confidence interval:
0.1007793 0.2032735
sample estimates:
p
0.145
10

R Commands for MATH 143 Examples of usage
pchisq(), qchisq(), chisq.test()
> 1 - pchisq(3.1309, 5) # gives P-value associated with X-squared stat 3.1309 when df=5
[1] 0.679813
> pchisq(3.1309, df=5, lower.tail=F) # same as above
[1] 0.679813
> qchisq(c(.001,.005,.01,.025,.05,.95,.975,.99,.995,.999), 2) # gives critical values like Table A
[1] 0.002001001 0.010025084 0.020100672 0.050635616 0.102586589 5.991464547 7.377758908
[8] 9.210340372 10.596634733 13.815510558
> qchisq(c(.999,.995,.99,.975,.95,.05,.025,.01,.005,.001), 2, lower.tail=F) # same as above
[1] 0.002001001 0.010025084 0.020100672 0.050635616 0.102586589 5.991464547 7.377758908
[8] 9.210340372 10.596634733 13.815510558
> observedCounts = c(35, 27, 33, 40, 47, 51)
> claimedProbabilities = c(.13, .13, .14, .16, .24, .20)
> chisq.test(observedCounts, p=claimedProbabilities) # goodness-of-fit test, assumes df = n-1
Chi-squared test for given probabilities
data: observedCounts
X-squared = 3.1309, df = 5, p-value = 0.6798
addmargins()
> blood = read.csv("http://www.calvin.edu/~scofield/data/comma/blood.csv")
> t = table(blood$Rh, blood$type)
> addmargins(t) # to add both row/column totals
A AB B O Sum
Neg 6 1 2 7 16
Pos 34 3 9 38 84
Sum 40 4 11 45 100
> addmargins(t, 1) # to add only column totals
A AB B O
Neg 6 1 2 7
Pos 34 3 9 38
Sum 40 4 11 45
> addmargins(t, 2) # to add only row totals
A AB B O Sum
Neg 6 1 2 7 16
Pos 34 3 9 38 84
11

R Commands for MATH 143 Examples of usage
prop.table()
> smoke = matrix(c(51,43,22,92,28,21,68,22,9),ncol=3,byrow=TRUE)
> colnames(smoke) = c("High","Low","Middle")
> rownames(smoke) = c("current","former","never")
> smoke = as.table(smoke)
> smoke
High Low Middle
current 51 43 22
former 92 28 21
never 68 22 9
> summary(smoke)
Number of cases in table: 356
Number of factors: 2
Test for independence of all factors:
Chisq = 18.51, df = 4, p-value = 0.0009808
> prop.table(smoke)
High Low Middle
current 0.14325843 0.12078652 0.06179775
former 0.25842697 0.07865169 0.05898876
never 0.19101124 0.06179775 0.02528090
> prop.table(smoke, 1)
High Low Middle
current 0.4396552 0.3706897 0.1896552
former 0.6524823 0.1985816 0.1489362
never 0.6868687 0.2222222 0.0909091
> barplot(smoke,legend=T,beside=T,main='Smoking Status by SES')
High Low Middle
current
former
never
Smoking Status by SES
0 20 40 60 80
12
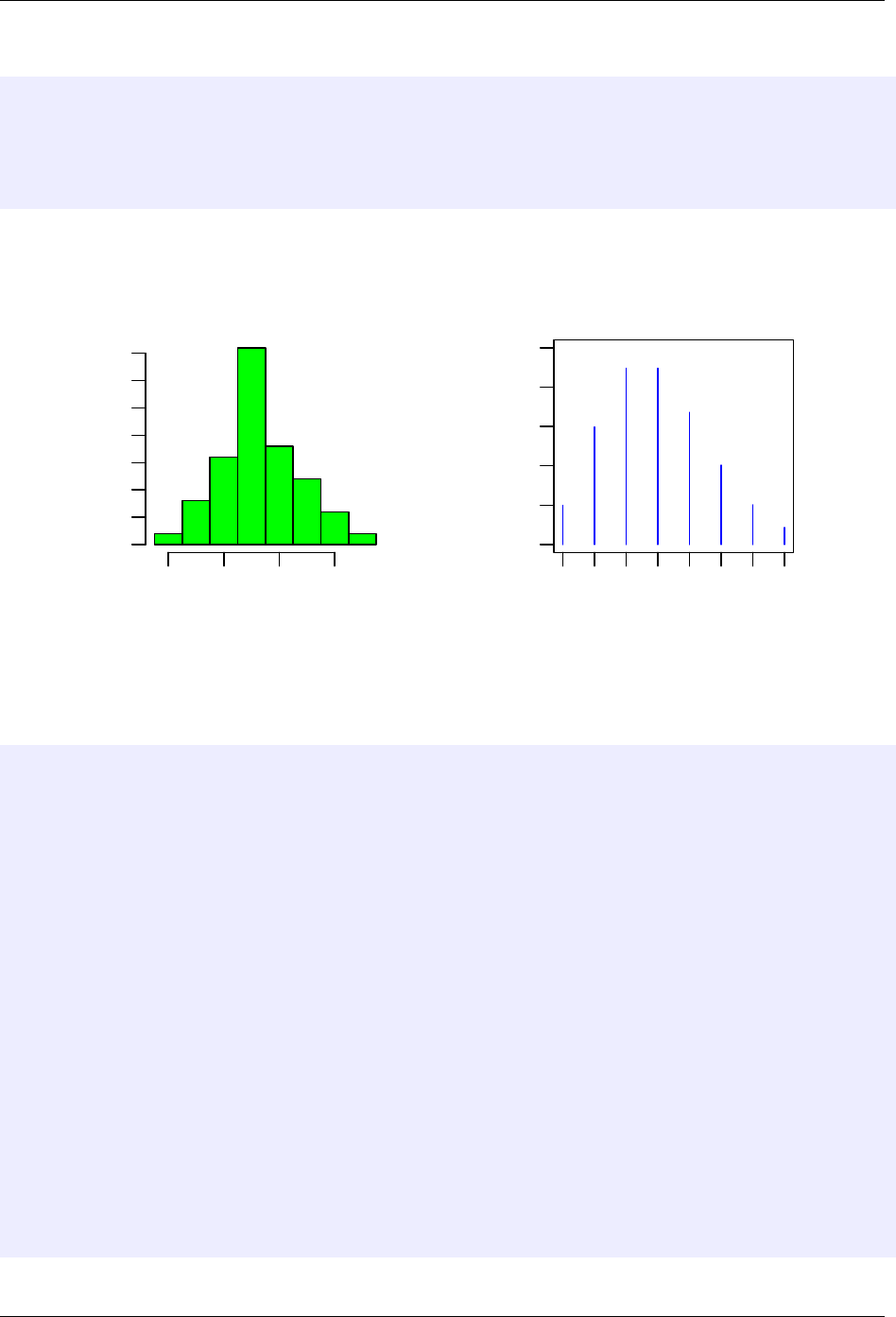
R Commands for MATH 143 Examples of usage
par()
> par(mfrow = c(1,2)) # set figure so next two plots appear side-by-side
> poisSamp = rpois(50, 3) # Draw sample of size 50 from Pois(3)
> maxX = max(poisSamp) # will help in setting horizontal plotting region
> hist(poisSamp, freq=F, breaks=-.5:(maxX+.5), col="green", xlab="Sampled values")
> plot(0:maxX, dpois(0:maxX, 3), type="h", ylim=c(0,.25), col="blue", main="Probabilities for Pois(3)")
Histogram of poisSamp
Sampled values
Density
0 2 4 6
0.00 0.15 0.30
0 1 2 3 4 5 6 7
0.00 0.10 0.20
Probabilities for Pois(3)
0:maxX
dpois(0:maxX, 3)
fisher.test()
> blood = read.csv("http://www.calvin.edu/~scofield/data/comma/blood.csv")
> tblood = xtabs(~Rh + type, data=blood)
> tblood # contingency table for blood type and Rh factor
type
Rh A AB B O
Neg 6 1 2 7
Pos 34 3 9 38
> chisq.test(tblood)
Pearson's Chi-squared test
data: tblood
X-squared = 0.3164, df = 3, p-value = 0.957
> fisher.test(tblood)
Fisher's Exact Test for Count Data
data: tblood
p-value = 0.8702
alternative hypothesis: two.sided
13

R Commands for MATH 143 Examples of usage
dpois(), ppois()
> dpois(2:7, 4.2) # probabilities of 2, 3, 4, 5, 6 or 7 successes in Pois(4.211)
[1] 0.13226099 0.18516538 0.19442365 0.16331587 0.11432111 0.06859266
> ppois(1, 4.2) # probability of 1 or fewer successes in Pois(4.2); same as sum(dpois(0:1, 4.2))
[1] 0.077977
> 1 - ppois(7, 4.2) # probability of 8 or more successes in Pois(4.2)
[1] 0.06394334
pnorm() qnorm(), rnorm(), dnorm()
> pnorm(17, 19, 3) # gives Prob[X < 17], when X ~ Norm(19, 3)
[1] 0.2524925
> qnorm(c(.95, .975, .995)) # obtain z* critical values for 90, 95, 99% CIs
[1] 1.644854 1.959964 2.575829
> nSamp = rnorm(10000, 7, 1.5) # draw random sample from Norm(7, 1.5)
> hist(nSamp, freq=F, col="green", main="Sampled values and population density curve")
> xs = seq(2, 12, .05)
> lines(xs, dnorm(xs, 7, 1.5), lwd=2, col="blue")
Sampled values and population density curve
nSamp
Density
2 4 6 8 10 12
0.00 0.10 0.20
14

R Commands for MATH 143 Examples of usage
qt(), pt(), rt(), dt()
> qt(c(.95, .975, .995), df=9) # critical values for 90, 95, 99% CIs for means
[1] 1.833113 2.262157 3.249836
> pt(-2.1, 11) # gives Prob[T < -2.1] when df = 11
[1] 0.02980016
> tSamp = rt(50, 11) # takes random sample of size 50 from t-dist with 11 dfs
> # code for comparing several t distributions to standard normal distribution
> xs = seq(-5,5,.01)
> plot(xs, dnorm(xs), type="l", lwd=2, col="black", ylab="pdf values",
+ main="Some t dists alongside standard normal curve")
> lines(xs, dt(xs, 1), lwd=2, col="blue")
> lines(xs, dt(xs, 4), lwd=2, col="red")
> lines(xs, dt(xs, 10), lwd=2, col="green")
> legend("topright",col=c("black","blue","red","green"),
+ legend=c("std. normal","t, df=1","t, df=4","t, df=10"), lty=1)
−4 −2 0 2 4
0.0 0.2 0.4
Some t dists alongside standard normal curve
xs
pdf values
std. normal
t, df=1
t, df=4
t, df=10
by()
> data(warpbreaks)
> by(warpbreaks$breaks, warpbreaks$tension, mean)
warpbreaks$tension: L
[1] 36.38889
---------------------------------------------------------------------------
warpbreaks$tension: M
[1] 26.38889
---------------------------------------------------------------------------
warpbreaks$tension: H
[1] 21.66667
15

R Commands for MATH 143 Examples of usage
t.test()
> data(sleep)
> t.test(extra ~ group, data=sleep) # 2-sample t with group id column
Welch Two Sample t-test
data: extra by group
t = -1.8608, df = 17.776, p-value = 0.0794
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-3.3654832 0.2054832
sample estimates:
mean in group 1 mean in group 2
0.75 2.33
> sleepGrp1 = sleep$extra[sleep$group==1]
> sleepGrp2 = sleep$extra[sleep$group==2]
> t.test(sleepGrp1, sleepGrp2, conf.level=.99) # 2-sample t, data in separate vectors
Welch Two Sample t-test
data: sleepGrp1 and sleepGrp2
t = -1.8608, df = 17.776, p-value = 0.0794
alternative hypothesis: true difference in means is not equal to 0
99 percent confidence interval:
-4.027633 0.867633
sample estimates:
mean of x mean of y
0.75 2.33
qqnorm(), qqline()
> qqnorm(precip, ylab = "Precipitation [in/yr] for 70 US cities", pch=19, cex=.6)
> qqline(precip) # Is this line helpful? Is it the one you would eyeball?
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
−2 −1 0 1 2
10 30 50
Normal Q−Q Plot
Theoretical Quantiles
Precipitation [in/yr] for 70 US cities
16

R Commands for MATH 143 Examples of usage
power.t.test()
> power.t.test(n=20, delta=.1, sd=.4, sig.level=.05) # tells how much power at these settings
Two-sample t test power calculation
n = 20
delta = 0.1
sd = 0.4
sig.level = 0.05
power = 0.1171781
alternative = two.sided
NOTE: n is number in *each* group
> power.t.test(delta=.1, sd=.4, sig.level=.05, power=.8) # tells sample size needed for desired power
Two-sample t test power calculation
n = 252.1281
delta = 0.1
sd = 0.4
sig.level = 0.05
power = 0.8
alternative = two.sided
NOTE: n is number in *each* group
anova()
require(lattice)
require(abd)
data(JetLagKnees)
xyplot(shift ~ treatment, JetLagKnees, type=c('p','a'), col="navy", pch=19, cex=.5)
anova( lm( shift ~ treatment, JetLagKnees ) )
Analysis of Variance Table
Response: shift
Df Sum Sq Mean Sq F value Pr(>F)
treatment 2 7.2245 3.6122 7.2894 0.004472 **
Residuals 19 9.4153 0.4955
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
treatment
shift
−2
−1
0
control eyes knee
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
17
